Recombinant Human Lysine-specific histone demethylase 1A (KDM1A), partial
-
中文名稱:人KDM1A重組蛋白
-
貨號:CSB-YP525314HU
-
規(guī)格:
-
來源:Yeast
-
其他:
-
中文名稱:人KDM1A重組蛋白
-
貨號:CSB-EP525314HU
-
規(guī)格:
-
來源:E.coli
-
其他:
-
中文名稱:人KDM1A重組蛋白
-
貨號:CSB-EP525314HU-B
-
規(guī)格:
-
來源:E.coli
-
共軛:Avi-tag Biotinylated
E. coli biotin ligase (BirA) is highly specific in covalently attaching biotin to the 15 amino acid AviTag peptide. This recombinant protein was biotinylated in vivo by AviTag-BirA technology, which method is BriA catalyzes amide linkage between the biotin and the specific lysine of the AviTag.
-
其他:
-
中文名稱:人KDM1A重組蛋白
-
貨號:CSB-BP525314HU
-
規(guī)格:
-
來源:Baculovirus
-
其他:
-
中文名稱:人KDM1A重組蛋白
-
貨號:CSB-MP525314HU
-
規(guī)格:
-
來源:Mammalian cell
-
其他:
產(chǎn)品詳情
-
純度:>85% (SDS-PAGE)
-
基因名:
-
Uniprot No.:
-
別名:Amine oxidase (flavin containing) domain 2; Amine oxidase, flavin containing, 2; AOF2; BHC110 ; BRAF35 HDAC complex protein BHC110; BRAF35-HDAC complex protein BHC110; BRAF35/HDAC complex, 110-kD subunit; CPRF; EC1; FAD binding protein BRAF35 HDAC complex, 110 kDa subunit; Flavin-containing amine oxidase domain-containing protein 2; KDM 1; KDM1; Kdm1a; KDM1A_HUMAN; KIAA0601; LSD 1; LSD1; Lysine (K) specific demethylase 1; Lysine (K) specific demethylase 1A; Lysine demethylase 1A; Lysine specific histone demethylase 1; Lysine specific histone demethylase 1A; Lysine-specific demethylase 1; Lysine-specific demethylase 1A; Lysine-specific histone demethylase 1A
-
種屬:Homo sapiens (Human)
-
蛋白長度:Partial
-
蛋白標(biāo)簽:Tag?type?will?be?determined?during?the?manufacturing?process.
The tag type will be determined during production process. If you have specified tag type, please tell us and we will develop the specified tag preferentially. -
產(chǎn)品提供形式:Lyophilized powder
Note: We will preferentially ship the format that we have in stock, however, if you have any special requirement for the format, please remark your requirement when placing the order, we will prepare according to your demand. -
復(fù)溶:We recommend that this vial be briefly centrifuged prior to opening to bring the contents to the bottom. Please reconstitute protein in deionized sterile water to a concentration of 0.1-1.0 mg/mL.We recommend to add 5-50% of glycerol (final concentration) and aliquot for long-term storage at -20℃/-80℃. Our default final concentration of glycerol is 50%. Customers could use it as reference.
-
儲存條件:Store at -20°C/-80°C upon receipt, aliquoting is necessary for mutiple use. Avoid repeated freeze-thaw cycles.
-
保質(zhì)期:The shelf life is related to many factors, storage state, buffer ingredients, storage temperature and the stability of the protein itself.
Generally, the shelf life of liquid form is 6 months at -20°C/-80°C. The shelf life of lyophilized form is 12 months at -20°C/-80°C. -
貨期:Delivery time may differ from different purchasing way or location, please kindly consult your local distributors for specific delivery time.Note: All of our proteins are default shipped with normal blue ice packs, if you request to ship with dry ice, please communicate with us in advance and extra fees will be charged.
-
注意事項:Repeated freezing and thawing is not recommended. Store working aliquots at 4°C for up to one week.
-
Datasheet :Please contact us to get it.
相關(guān)產(chǎn)品
靶點(diǎn)詳情
-
功能:Histone demethylase that can demethylate both 'Lys-4' (H3K4me) and 'Lys-9' (H3K9me) of histone H3, thereby acting as a coactivator or a corepressor, depending on the context. Acts by oxidizing the substrate by FAD to generate the corresponding imine that is subsequently hydrolyzed. Acts as a corepressor by mediating demethylation of H3K4me, a specific tag for epigenetic transcriptional activation. Demethylates both mono- (H3K4me1) and di-methylated (H3K4me2) H3K4me. May play a role in the repression of neuronal genes. Alone, it is unable to demethylate H3K4me on nucleosomes and requires the presence of RCOR1/CoREST to achieve such activity. Also acts as a coactivator of androgen receptor (AR)-dependent transcription, by being recruited to AR target genes and mediating demethylation of H3K9me, a specific tag for epigenetic transcriptional repression. The presence of PRKCB in AR-containing complexes, which mediates phosphorylation of 'Thr-6' of histone H3 (H3T6ph), a specific tag that prevents demethylation H3K4me, prevents H3K4me demethylase activity of KDM1A. Demethylates di-methylated 'Lys-370' of p53/TP53 which prevents interaction of p53/TP53 with TP53BP1 and represses p53/TP53-mediated transcriptional activation. Demethylates and stabilizes the DNA methylase DNMT1. Required for gastrulation during embryogenesis. Component of a RCOR/GFI/KDM1A/HDAC complex that suppresses, via histone deacetylase (HDAC) recruitment, a number of genes implicated in multilineage blood cell development. Effector of SNAI1-mediated transcription repression of E-cadherin/CDH1, CDN7 and KRT8. Required for the maintenance of the silenced state of the SNAI1 target genes E-cadherin/CDH1 and CDN7.
-
基因功能參考文獻(xiàn):
- the role of the LSD1 complex in the reactivation of HIV-1 transcription from latency PMID: 30105631
- LSD1 and JMJD2C disable oncogenic Ras- or Braf-induced senescence by enabling the expression of E2F target genes PMID: 29438700
- LSD1 inhibition may serve as a promising epigenetic adjuvant therapy to subvert its pleiotropic roles in breast cancer progression and treatment resistance. PMID: 29311580
- Results show that KDM1A is upregulated in the gastric cancer (GC) cells and tissues. Its expression is negatively modulated by mir329 which directly targets its 3'utr. PMID: 29130516
- FOXP4-AS1 is overexpressed in osteosarcoma (OS), and is the independent risk factor in OS prognosis. Upregulated FOXP4-AS1 promotes the proliferation, migration and cell cycle, but inhibits apoptosis of OS cells. Furthermore, FOXP4-AS1 participates in the development and progression of OS by downregulating LATS1 via binding to LSD1 and EZH2. PMID: 29859193
- Enhancer of Zeste Homolog 2 (EZH2), SET domain, bifurcated 1 protein (SETDB1), lysine-specific histone demethylase 1 (LSD1), histone H3 methylation (H3K9me3 and H3K27me3) expression are altered in colorectal cancer (CRC) and may play a role in colorectal carcinogenesis. PMID: 30105513
- the crystal structure of LSD1 in complex with the inhibitor 4-[5-(Piperidin-4-ylmethoxy)-2-(p-tolyl)pyridin-3-yl]benzonitrile at 2.96 A, is reported. PMID: 29949906
- This study showed that tetrahydrofolate stably binds to the LSD1/CoREST complex, in its open conformation, at its entrance. PMID: 29161028
- Study reveals LSD1 as a molecular rheostat selectively regulating H3K9 demethylation at cell cycle gene loci, thereby representing a key player in oncogenesis and a viable target for cancer therapy. PMID: 28991226
- KDM1A is a unique epigenetic modifier with the ability to maintain interactions with a variety of different protein complexes, noncoding RNAs, microRNAs, and transcription factors. The functional significance of KDM1A is maintained by its interactions at multiple sites in the genome, particularly its binding to promoters and enhancers. [review] PMID: 29921310
- our findings identified LSD1 as a novel negative regulator of autophagy through the mTOR signaling pathway in ovarian cancer HO8910 cells and indicated that LSD1 may function as a driving factor of ovarian cancer progression via deregulating autophagy. PMID: 29749504
- a small-molecule LSD1 inhibitor-SP-2509-blocks important demethylase-independent functions and suppresses castration-resistant prostate cancer cell viability demonstrates the potential of LSD1 inhibition in this disease. PMID: 29581250
- expression of the nearby cyclindependent kinase inhibitor 1C (CDKN1C) gene was revealed to be upregulated after SP3 knockdown in cells that possessed non-risk alleles. This suggests that CDKN1C is potentially one of the functional targets of SNP rs163184, which modulates the binding activity of the locus for Sp3 and Lsd1/Kdm1a PMID: 29207083
- Silencing of LSD1 is capable of removing the mono- and dimethyl groups from H3K4, and upregulating the histone acetylation of H3 in JeKo1 and MOLT4 cells. The silencing of LSD1 inhibited cell growth and induced cell apoptosis. PMID: 28627608
- LSD1 and ERRalpha coregulate several target genes involved in cell migration, including the MMP1 matrix metallo-protease, also activated through H3K9 demethylation at the transcriptional start site. PMID: 28348226
- concomitant LSD1 and HDAC inhibition synergistically induces cell death in rhabdomyosarcoma cells by shifting the ratio of pro- and antiapoptotic BCL-2 proteins in favor of apoptosis, thereby engaging the intrinsic apoptotic pathway PMID: 28617441
- Results show that LSD1 protein level is elevated in prostate tumors (PCa) and correlate with faster tumor growth in xenograft mouse models. Knockdown of LSD1 reduces PCa cell viability, and gene expression data suggest a dual coregulatory role of LSD1 for VDR, acting as a coactivator and corepressor in a locus-specific manner. PMID: 28811844
- Study showed that repressed HBV cccDNA chromatin state is activated by LSD1 by demethylating H3K9. Also, LSD1 was shown to be recruited to viral promoters in an HBx dependent manner. PMID: 27174370
- LSD1 knockdown has a broad effect on histone lysine methylation. PMID: 28601046
- analysis of a novel cellular stress response mechanism in cancer cells and a key role of LSD1/SIRT1/KU70 dynamic interaction in regulating DNA repair and mutation acquisition PMID: 27384990
- Patients with mutations 6 showed higher rate of achieving major molecular response than those<6 (P=0.0381). Mutations in epigenetic regulator, ASXL1, TET2, TET3, KDM1A and MSH6 were found in 25% of patients. TET2 or TET3, AKT1 and RUNX1 were mutated in one patient each. ASXL1 was mutated within exon 12 in three cases PMID: 28452984
- Study demonstrated that LSD1 in cooperation with MYCN controls cell migration and invasiveness of neuroblastoma cells through transcription regulation of the metastatic suppressor NDRG1. PMID: 27894074
- Results show that LSD1 could directly bind to the promoter of P21, inducing H3K4me2 demethylation. PMID: 28209170
- The autophagy induced by 2-PCPA requires LC3-II processing machinery..2-PCPA treatment induces the change of global gene expression program, including a series of autophagy-related genes, such as SQSTM1/p62. Taken together, our data indicate that KDM1A/LSD1 inhibitors induce autophagy through affecting the expression of autophagy-related genes and in a BECN1-independent manner PMID: 28800922
- LSD1 is specifically mislocalized to pathological protein aggregates in the Alzheimer's disease and frontotemporal dementia patients brain. PMID: 28993646
- these results demonstrate crosstalk between the lysine demethylase KDM1A and the DNA methyltransferase DNMT1, which could be involved in carcinogenesis independently of its role in DNA methylation PMID: 27449289
- LSD1 inhibition with HCI-2509 decreases the c-MYC level in poorly differentiated prostate cancer cell lines. PMID: 27349498
- Such a relatively simple strategy uncovered two new classes of LSD1 inhibitors , which reveal unexpected binding modes at the same protein surface, establishing specific and common contacts, and highlight novel routes for the development of compounds targeting epigenetic processes. PMID: 27626075
- we show that KDM1A promotes cancer metastasis in non-small cell lung cancer cells by repressing TIMP3 (tissue inhibitor of metalloproteinase 3) expression. PMID: 27058897
- Results show that LSD1 expression was upregulated in breast carcinoma and, induced EMT in breast cancer cells through demethylation of H3K4me at the E-cadherin promoter. Its phosphorylation at Ser112 is crucial for these functions. PMID: 27572339
- LSD1 directly associates with the promoter of the HEYL gene. PMID: 27018646
- LSD1 controls autophagy in neuroblastoma cells through SESN2 transcription regulation. PMID: 28783174
- Mechanistic investigations showed that LINC01133 could interact with EZH2, LSD1 and recruit them to KLF2, P21 or E-cadherin promoter regions to repress their transcription. PMID: 26840083
- estrogen induced TFPI-2 expression in MCF7 cells is mediated by ERalpha and also by the action of LSD1. PMID: 28088469
- CENPE was regulated by the co-binding of LSD1 and AR to its promoter, which was associated with loss of RB1 in CRPC. PMID: 28916652
- RBPJ interacts with L3MBTL3 to promote repression of Notch signaling via histone demethylase KDM1A. PMID: 29030483
- High LSD1 expression is associated with small-cell lung cancer. PMID: 28667074
- The role of LSD1 in the EMT process. PMID: 28720390
- p53-CTD inhibits LSD1 activity and that the direct association between the two proteins can contribute to their functional cross-talk. PMID: 28784588
- LSD1 is a key regulator of HIF1alpha/VEGF-mediated tumor angiogenesis by antagonizing the crosstalk between PTMs involving HIF1alpha protein degradation. PMID: 28534506
- This study identified histone modification by SUMO as the first post-translational modification that stimulates intranucleosomal demethylation by the developmentally critical LSD1-CoREST complex. PMID: 28832116
- These findings provide insight into the regulation of LSD1 and Epithelial-to-Mesenchymal Transition (EMT) and identify MOF as a critical suppressor of EMT and tumor progression. PMID: 27292636
- High LSD1 expression is associated with colon cancer. PMID: 28121627
- results demonstrate that CoREST and LSD1 downregulate the Notch pathway in the developing cerebral cortex PMID: 27112428
- LSD1 is a primary and positive regulator of the HPV16 E7-induced epithelial-mesenchymal transition in cervical cancer PMID: 27894088
- KDM1A inhibitors may be utilized to alleviate acquired resistance to sorafenib in hepatocellular carcinoma cells. PMID: 28377178
- This study identified the lysine-specific demethylase KDM1A as a novel interaction partner of ZEB2 and demonstrated that mouse and human T-cell acute lymphoblastic leukemia cells with increased ZEB2 levels critically depend on KDM1A activity for survival. PMID: 28069602
- LSD1 overexpression is associated with non-small cell lung cancer. PMID: 28223039
- a model in which the EZH2/HOXA11-AS/LSD1 complex and HOXA11-AS/miR-1297/EZH2 cross-talk serve as critical effectors in gastric cancer tumorigenesis and progression. PMID: 27651312
- these results demonstrate the relevance of LSD1 in gene regulation and how even moderate alterations in its stability, catalytic activity and binding properties can strongly affect organism development. PMID: 27094131
顯示更多
收起更多
-
相關(guān)疾?。?/div>Cleft palate, psychomotor retardation, and distinctive facial features (CPRF)亞細(xì)胞定位:Nucleus.蛋白家族:Flavin monoamine oxidase family組織特異性:Ubiquitously expressed.數(shù)據(jù)庫鏈接:
Most popular with customers
-
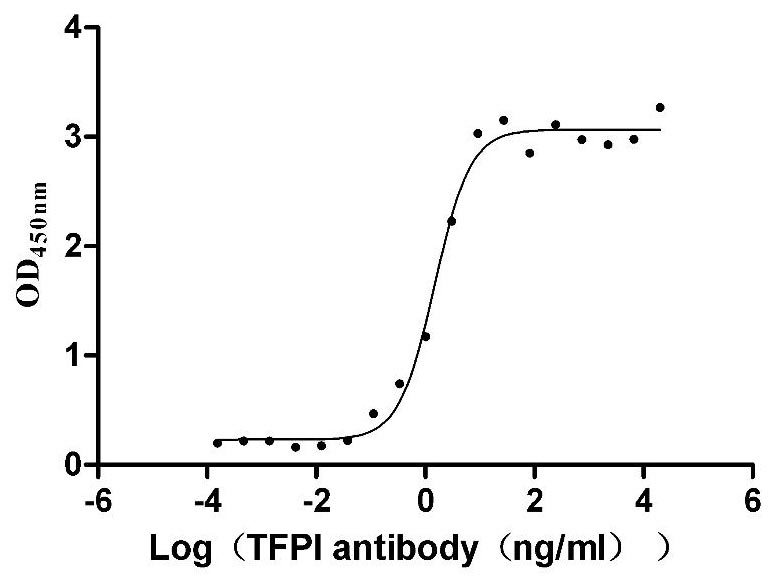
Recombinant Human Tissue factor pathway inhibitor (TFPI), partial (Active)
Express system: Mammalian cell
Species: Homo sapiens (Human)
-
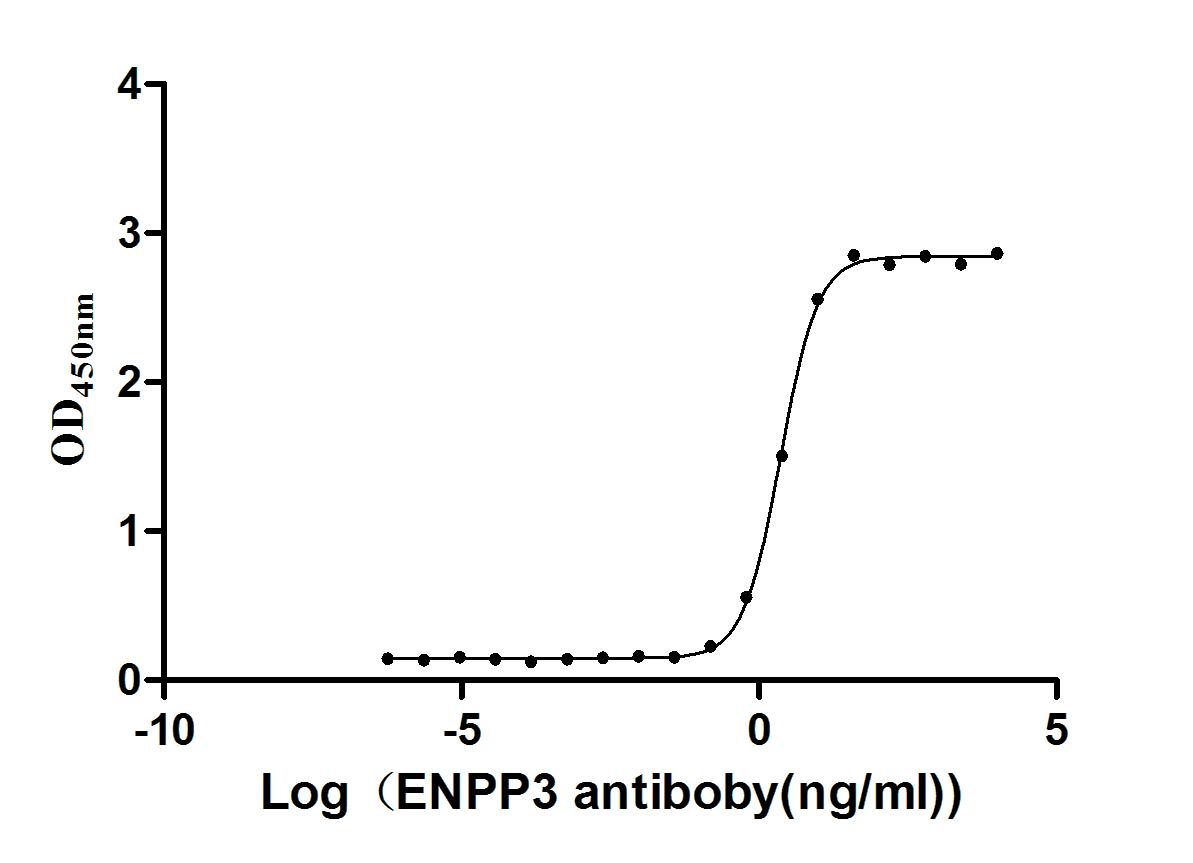
Express system: Mammalian cell
Species: Homo sapiens (Human)
-
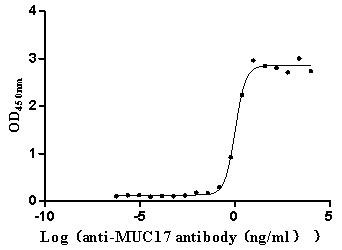
Recombinant Human Mucin-17 (MUC17), partial (Active)
Express system: Mammalian cell
Species: Homo sapiens (Human)
-
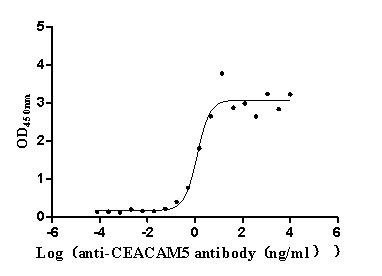
Express system: Mammalian cell
Species: Homo sapiens (Human)
-
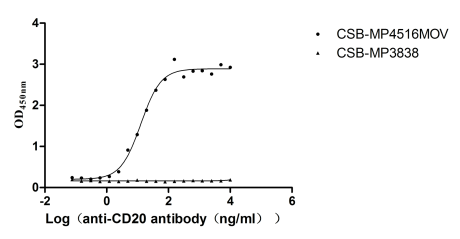
Recombinant Macaca fascicularis Membrane spanning 4-domains A1 (MS4A1)-VLPs (Active)
Express system: Mammalian cell
Species: Macaca fascicularis (Crab-eating macaque) (Cynomolgus monkey)
-
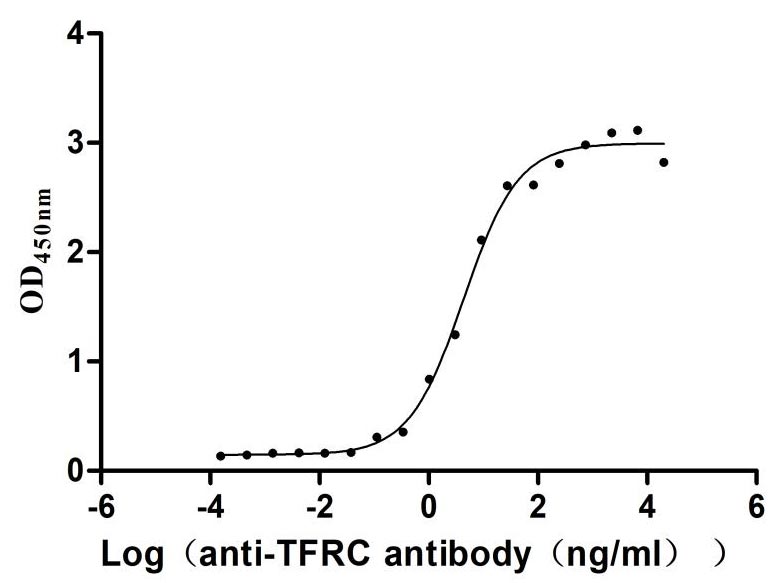
Recombinant Human Transferrin receptor protein 1 (TFRC), partial (Active)
Express system: Mammalian cell
Species: Homo sapiens (Human)
-
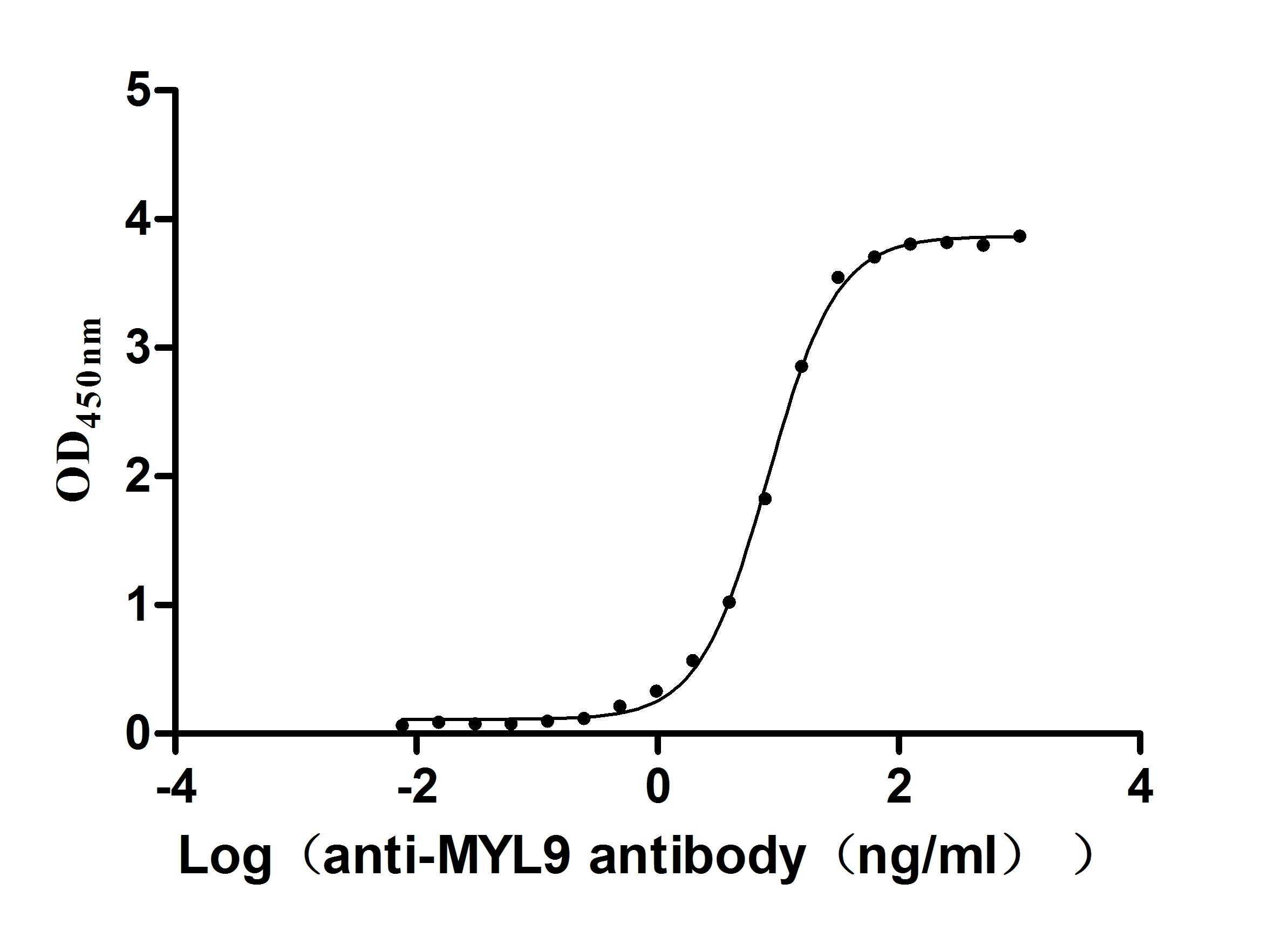
Recombinant Human Myosin regulatory light chain 12B(MYL12B) (Active)
Express system: E.coli
Species: Homo sapiens (Human)
-
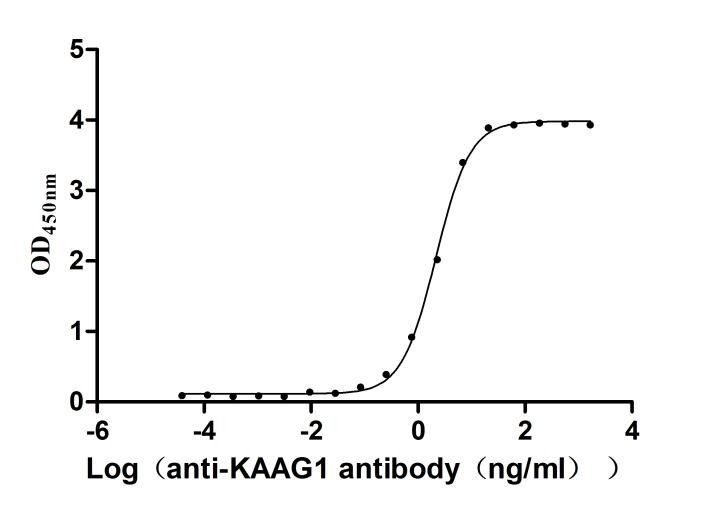
Recombinant Human Kidney-associated antigen 1(KAAG1) (Active)
Express system: E.coli
Species: Homo sapiens (Human)